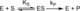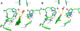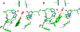Por favor, use este identificador para citar o enlazar este ítem:
https://doi.org/10.3390/ijms25136891 Twittear
Twittear

| Título: | Molecular docking studies of ortho-substituted phenols to tyrosinase helps discern if a molecule can be an enzyme substrate |
| Fecha de publicación: | 23-jun-2024 |
| Fecha de defensa / creación: | 20-may-2024 |
| Editorial: | MDPI |
| Cita bibliográfica: | International Journal of Molecular Sciences 2024, 25, 6891. ISSN 1661-6596 |
| ISSN: | 1661-6596 |
| Materias relacionadas: | CDU::5 - Ciencias puras y naturales::57 - Biología::577 - Bioquímica. Biología molecular. Biofísica |
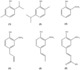
| Palabras clave: | Tyrosinase Substrates Inhibitors Ortho-substituted phenols Molecular docking |
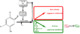
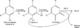
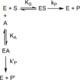
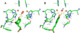
| Resumen: | Phenolic compounds with a position ortho to the free phenolic hydroxyl group occupied can be tyrosinase substrates. However, ortho-substituted compounds are usually described as inhibitors. The mechanism of action of tyrosinase on monophenols is complex, and if they are ortho-substituted, it is more complicated. It can be shown that many of these molecules can become substrates of the enzyme in the presence of catalytic o-diphenol, MBTH, or in the presence of hydrogen peroxide. Docking studies can help discern whether a molecule can behave as a substrate or inhibitor of the enzyme. Specifically, phenols such as thymol, carvacrol, guaiacol, eugenol, isoeugenol, and ferulic acid are substrates of tyrosinase, and docking simulations to the active center of the enzyme predict this since the distance of the peroxide oxygen from the oxy-tyrosinase form to the ortho position of the phenolic hydroxyl is adequate for the electrophilic attack reaction that gives rise to hydroxylation occurring. |
| Autor/es principal/es: | Montenegro-Arce, María Fernanda Teruel-Puche, Jose Antonio Garcia-Molina, Pablo Tudela-Serrano, Jose Rodriguez-Lopez, Jose Neptuno Garcia-Canovas, Francisco Garcia-Molina, Francisco |
| Versión del editor: | https://www.mdpi.com/1422-0067/25/13/6891 |
| URI: | http://hdl.handle.net/10201/153521 |
| DOI: | https://doi.org/10.3390/ijms25136891 |
| Tipo de documento: | info:eu-repo/semantics/dataset |
| Número páginas / Extensión: | 20 |
| Derechos: | info:eu-repo/semantics/openAccess Attribution-NonCommercial-NoDerivatives 4.0 Internacional |
| Aparece en las colecciones: | Datos de investigación |
Ficheros en este ítem:
Este ítem está sujeto a una licencia Creative Commons Licencia Creative Commons